Research Computing Teams #114, 18 Mar 2022
Hi!
It’s been pointed out I have my view of the role of specialization in research computing teams seems a bit contradictory. I am very pro-specialization, but anti-siloing. Fair point! Let’s dig more into that.
No research computing and data team can do everything. It’s not possible. Pretending otherwise leads to unhappy team members and unsatisfied researchers. And as I like to point out, there’s already tonnes of computing things our teams don’t do for researchers. We don’t help them with their printers, or lay out designs of posters for conferences. So my advice is wherever possible — and it’s more possible than you think — is to add a few things to that list. To niche down into a specialization.
The most meaningful specialization is of the form of “we help [community] with [problem]”. One very effective way to do that is where community is a particular discipline (defined broadly). “We help wet lab molecular biologists with their bioinformatics analyses”. “We help social sciences researchers with their GIS software development needs”. “We help researchers with long term data retention needs affordably and reliably store and share their data”. “We help health scientists write and deploy desktop and mobile data collection applications”. “We help researchers new to quantitative methods with data science”. “We help researchers with web applications deploy, operate and maintain their application stacks”.
There are a lot of advantages of such a specialization:
- It lets your team get stronger, faster. Team members can go very deep into both understanding particular community needs and the details of a problem domain, building their skills quickly.
- It lets your team get more efficient, as your teams build components of solutions that can be reused.
- It helps you recruit researcher clients more easily, because it’s very easy for them to understand what you can help with, and to recommend you to others in their community.
- It helps you recruit the right kind of staff more easily, because it’s very easy for them to understand what you do and why they’d want to work there.
- It helps you communicate your value to decision makers in your institutions (or amongst your funders) and demonstrate alignment with their priorities.
A decent sized team could potentially have two or three of those specializations, aligned either along the community or problem axes, and still be sharply focussed. It starts to look like a core facility, which faculty, institutional decision makers, and funders all instinctively understand.
A significantly weaker and less meaningful form of specialization is, unfortunately, a little easier to fall into. It’s more inward looking and input based rather than focused on researchers and their needs. It’s to lean on the tools you use. To focus shallowly on the activities you perform. “We write C++ code for research”. “We run a cluster researchers can use”.
There are three big problems with this approach.
The first is with communicating your existence to researchers at all, much less attracting them as clients. Few researchers have a problem they think of as primarily a C++ problem, or a running-cluster problem. By and large, they have (say) functionality they want to add to a groundwater data analysis code, or the need to run an ensemble of simulation-analysis pipelines. Maybe C++ or a particular kind of cluster is one of the tools that can be used to solve their problem, and if so, great! But there are likely other options they could be using, too. (Do you doubt me about finding researchers? Because I’ve been on several function-specific teams which were “the only game in town” at particular institutions, and we were constantly shocked at how many researchers had just never heard of us.)
The second is in having those researchers value your work. If you provide undifferentiated C++ programming or cluster administration, you’ll always do a worse job for that particular researcher than a team that did software development specifically for subsurface science, or a computing resource that can queue complex simulation-analysis-pipeline workloads. There will always be researchers who can take advantage of the undifferentiated offering, but they’ll be the ones who could just as easily move elsewhere the moment the opportunity arose. They won’t develop any particular relationship with the team nor advocate strongly for the team in particular.
The third is communicating your value to decision makers, funders, or potential staff members. “We write C++ research software”, “we run a big cluster” isn’t a compelling story. If you back it up with undifferentiated statistics - which is all you can really offer - even less so. “We completed 15 projects last year!”. So? Is that a lot? A little? How did that move a field forward? “We run at 91% utilization!”. Is that good? Bad? How did that help the University? You can try to beat the drum for case studies and high-impact publications resulting from the inputs. But it’s always going to be an uphill battle trying to get research groups to write them up or even bother notifying you about their paper. It’s hard because you’re running an undifferentiated resource, one they could replace if necessary. Writing something up for you is just one more low-priority item for the todo list.
Still, there’s a reason why this approach is common amongst teams to organize around - we’ll come back to it in a moment.
Because there’s an even worse way to “specialize”, and I see worrying trends of it happening as countries and regions rightly start taking research computing and data more seriously. And that’s by “pillars” (read silos) - software, data, and infrastructure.
This approach kind of made sense in the 90s or early 2000s. Systems were getting increasingly homogenous, software was all of a certain kind (3d physical simulations) that ran on those systems. Data teams if they existed at all were separate entirely, focussed on different users, in business schools or social sciences, and were all SAS/SPSS/whatever and SQL. There wasn’t much overlap.
Historically teams tended to be broken up that way, and that’s the genesis of most of today’s research computing teams in Universities. “Because we’ve always done it that way” caries a lot of weight in old institutions. And you see their legacy still.
Today, if you were starting from scratch, you’d never organize teams around those functions. It’s absurd. Everything in our field today is a combination of the three facets of research computing and data. The “data collection tools for health researchers”, for instance, is a combination of infrastructure (deployment of a backend), data management (handling sensitive data), and software development (the application itself). Bioinformatics is inherently software + data.
The increasingly unified, cross-function nature of such work is a challenge even for your faithful correspondent. Take a look at the articles in the data management section this week, for instance - CI/CD pipelines for data artifacts. Is that an infra thing, a software thing, or data management? How about cloud-friendly libraries for geospatial data? How about data analysis toolkits in the browser? Which section should those go in
For organizing a newsletter roundup it can still be helpful to bin articles by which facet is most distinctive, and mis-binning is mostly harmless.
If you’re deciding on how to fund efforts, however, the stakes are much higher! In that case, you really need to consider everything holistically, and binning into silos is a terrible idea. Data without software is useless, as is software that’s not deployed anywhere, as is a compute infrastructure with no storage. Even in the 90s, the best software teams were very aware of the systems their tools would run on and coded accordingly, and the systems teams were very aware of their workloads. Having the teams communicate across silo boundaries was something the best teams did. Both sides needed to know what was implementable. Organizing funding by silo right from the beginning makes that cross-silo communication much harder.
We as human beings need to organize somehow, though - we can’t communicate with everyone else in research computing and data equally. The highest impact teams I’ve seen (and, not coincidentally, the ones that are least worried about their funding in the coming years) are those that have a specific focus, usually disciplinary (mirroring the research community focus they support), and have cross-functional expertise in their team. They have enough knowledge to help a researcher see a problem through beginning to end. The researchers see them as partners, and will advocate for them. And that’s because they’ve organized themselves around what they’re supporting, particular kinds of projects, rather than internally focussed on a tool they use.
Does that line up with what you see on the ground? Are their other ways of specializing or organizing that you’ve seen work? Email me with that or with any other comments or questions by hitting reply or sending an email to jonathan@researchcomputingteams.org.
With that, on to the roundup!
Managing Teams
The Uninspiring Manager - Matt Schellhas
Too much of what I imagined an excellent manager should be early in my career (not having really seen any) was focussed on personal characteristics. Such leaders should be engaging, people-persons… and inspiring.
It’s not that any of those things is bad, of course. Those are all traits and behaviours that can be put to good use as a manager or lead! But so can the default behaviours of quiet, attentive, carefully-thinks-before-speaking introverts. Or of keep-ticking-things-off-the-list achievers. Or of caring, emotionally intuitive, confidantes. Each has default behaviours that are extremely helpful in the right circumstance, and are downright liabilities in others.
Schellhas makes the case against needing to be inspiring to be a good manager. In his view, people mostly already want to do a good job, and what they need to accomplish that is: information; skill; and willpower.
Inspiration doesn’t help at all with the first two, and only temporarily for the third. A better use of energy than temporarily boosting willpower with some rah rah stuff, he says, would be to put effort into fixing willpower-sapping features of the job. Fixing flaky CI systems that make writing tests seem useless, or removing unnecessary reporting.
One thing Schellhas doesn’t call out that I’ll add - being able to communicate a clear vision for the team is, in my estimation, a requirement of the job. People need to know what they are aiming for, and how their work fits into that. And that vision might even be inspiring! But that inspiration best comes from the work and goals themselves, not from some charismatic leader.
Trunk and Branches Model for Scaling Infrastructure Organizations - Will Larson
Larson here is talking specifically about infrastructure teams. But this could apply to growing research computing and data teams.
The constraints are keeping the core operations of an initial team with a broad remit going while growing and adding capabilities. The model he recommends and has seen work is to grow slowly and, as particular well-defined areas start emerging as being important, to “bud off” a branch sub-team with that specialty while the “trunk” team continues on with reduced scope. In our context, this could be two officially different teams with different managers/leads, or, maybe less ideally, overlapping groups of competence within a single large team.
(By the way, this is all perfectly consistent with beginning with a team that already has a well-defined specialty - GIS data science or web/mobile apps for data collection or managing clusters for AI/ML workloads. The wonderful thing about research, and expertise in general, is that it’s fractal. No matter how small a chunk of the big picture you start with, as you zoom further in you see more and more complexity. You discover whole new parts of the map to explore and become knowledgeable in.)
The key things Larson emphasizes here is to be clear (internally and externally) about what who owns what responsibilities as a team branches off.
I really like this documented hiring process by 18F in the US Government. It’s a well thought out process, and it’s written in a way that you could send to candidates so they know exactly what to expect. It’s even in GitHub. I also really like their technical pre-work - it’s either to provide some code they’ve worked on, or to do one of four exercises. The exercises are simple but non-trivial get-and-process-data exercises that would give a lot more confidence about ability to do real programming tasks than some kind of fizz-buzz.
Managing Your Own Career
Minto Pyramid - Adam Amran, Untools
Amran gives a very clear formula here for emails that you also see in advice for briefing boards (or in our case, e.g., scientific advisory committee.). Start with a one-sentence paragraph of the conclusion (or the ask); then a listing of the key arguments; then the supporting details. I’d add that the subject line should reflect the conclusion/ask.
I’ve been thinking a lot about this recently. I’m generally ok about writing to-the-point, skimmable emails. But that skill may have atrophied a bit recently. In my previous job, I didn’t send a lot of emails (we used slack internally mostly). When I did send them, as a manager and as a big fish in a small pond, I… could kind of assume my emails would be carefully read. In retrospect I kind of got lazy and leaned on that, which I now feel sheepish about.
I’m in now an individual contributor in a very email-heavy organization. I can see very clearly the effects of getting lazy about sending bottom-line-up-front emails. I’ve tried to communicate some points via email to some very busy account managers in the past couple weeks, and some of the points just did not get across. So it’s time to up my game again.
Not every email lends itself to this formula, of course, but sticking to bottom-line-up-front is a good general principle.
Product Management and Working with Research Communities
Journal clubs, ranked from worst to best - Guillaume Filion
Journal clubs can be great. The usual failure mode is that few other than the presenter actually reads the paper in any depth, so there’s not much discussion afterwards.
Fillon ranks the journal club mechanisms he’s seen work the best.
- If the session will cover one paper, have everyone read the paper and on game day randomly assign explaining one figure to each of the participants. That way everyone is, um, “incentivized” to have read the paper ahead of time.
- Alternately, have each person cover one paper, curated from a list of thematically-linked papers. The discussion of each paper will necessarily be a little shallow, but this will let you really go over an area of research.
Cancer R&D funding decimated during Covid crisis, says NCRI - Sophie Inge, Research Professional News
There has been and will continue to be a huge influx of research money into health, but the past few years have been really rough on cancer reseach. Not only is it no longer anyone’s #1 health research priority, a lot of the research funds comes from charitable giving which the pandemic-related recession hit hard.
NCRI reports a drop of 9% of funding, which doesn’t sound like much but the majority of funding spending is on salaries. No doubt infectious disease research rose at least as much in absolute numbers, but people with years of training and expertise can’t be repurposed as easily as dollars can.
In general I think the coming years are going to see increasing changes from the pre-2020 baseline research funding landscape.
AI did not cover itself in glory over the past two years of COVID, and neither I’d argue did HPC - while bread-and-butter high-throughput computing and on-the-desktop bioinformatics workflows were invaluable.
Building a technical conceptual map (or “conceptual diagram”) of a subject. This is a very useful technique for learning a subject, or for preparing for teach a subject. It’s sort of a typed mind map. It can be used for very specific topics such as here Javascript’s reduce() method, or for the topic of a semester long course. Either way it’s very helpful for organizing an initially amorphous mass of concepts and identifying the key things to cover and in what order.
Cool Research Computing Projects
Black hole “billiards” may explain strange aspects of 2019 black hole merger - Jennifer Ouellette
Oulette at Ars does a great job describing a recent nature paper where the authors (Samsing et al) ran a large ensemble of 3d post-newtonian particle simulations trying to understand a weird merger, GW190521, that VIRGO/LIGO detected in 2019. The merger implied a total of 150 solar masses of black holes involved, which is a weird number - most black holes are either a few to a couple dozen times solar, or much more massive. Even weirder, a month later there was a big flash in the area.
In the paper by Samsing et al., the ensemble of simulations showed that adding a third black hole increases the odds of eccentric orbits by two orders of magnitude (and accounts for the observed strongly misaligned spin axes). If the merger occurred in an environment with multiple black holes in a big disk - say circulating a supermassive black hole - that could account for all observations.
Relatedly, if you want cool black hole pictures, here’s a blog post (including a figure below) demonstrating increasingly realistic rendering of accretion disks around black holes with the Unity game engine.

Research Software Development
Survey reveals 6000+ people develop and maintain vital research software for Australian research - Jo Savill, Australian Research Data Commons (ARDC)
Research Software Capability in Australia - Michelle Barker and Markus Buchhorn
Interesting results from a late-2021 ARDC survey on research software capability, of 70 managers of Australian research computing and data groups. Results were scaled to try to give an estimate of all-of-Australia numbers.
The article by Savill gives an overview, and the full report by Barker and Buchhorn is interesting reading. Some key findings taken from the article and the report:
- About 6,000 people (and about 2,500 FTEs) working in roles that provide software development.
- That’s about 1 FTE worth of software developer (broadly defined) effort per 40 researchers.
- 46% of respondents perceived that the skills of their research software capability they manage were adequate.
- 78% of respondents answered “yes” or “maybe” to a question on whether these personnel had access to mechanisms to improve their skills. (LJD: That’s not super encouraging. Free Coursera courses is a mechanism to improve skills…)
- 80 different job titles were listed as used for these staff. (LJD: 80!!!!)
- Only 33% of these staff have permanent employment.
- 56% of researchers did not feel there was adequate research software capability in their area - rising to 60% of those with a focus on an entire discipline and 75% for those with a whole-of-University focus. (LJD: These numbers are consistent with sub-disciplines that are heavy software users and probably contributors disproportionately returning surveys, which I think is what I’d expect.)
These numbers - and the problems they suggest - seem plausible to me for Canada, as well. The reliance on unfunded and part-time software development and maintenance is a real issue, as is the lack of any kind of coherent career track (80 job titles!!). The good news is that 43% of respondents had plans to recruit more people into those roles over the next 1-3 years.
Do these proportions seem about right in your neck of the woods (be that geography or discipline?). Do things look like they’re getting better or worse?
Building a Backlog Your Team Will Love - Blurbs By Amy
At my last job — and this was my fault — the backlog became an undifferentiated mass. We did keep higher priorities to the top, and lower priority items to the bottom. There was a lot there, though, some we were clearly never going to get to, and the tickets were of very different size… yeah, it wasn’t great.
Here Amy outlines her teams workflow, and it looks pretty good:
- An inbox, called “Add New Here” to make it completely unambiguous that new tasks go there.
- Have “Not planning to do” and “Future Nice to Haves” sections for record-keeping but to keep these from clogging up the works.
- The workflow is that inbox items get groomed and then immediately triaged (duaged?) into “Groomed + Prioritized” for high-priority tasks, and “Groomed + Not Prioritized” for the lower-priority tasks, or one of the parking lots.
- Then tasks get selected into Next Sprint tasks.
I like the unambiguous process here. You’re clearly doing something wrong if you add a ticket anywhere than “add new here”, for instance. Having two different parking lot sections seems a bit much, but it lets you move tasks into “Future nice to haves” without feeling like you’re consigning it to the dustbin. Over time clean-up can move things into Not Planning to Do, and adjust the priorities of groomed tasks.
Congratulations to the Python core team for deprecating and then removing some old stdlib modules. Choosing to stop supporting something is genuinely hard, and at Python scales even clearly past-its-best-before-date modules like cgi (cgi!!!) are probably used by thousands of people. Getting rid of a stdlib (and thus de facto default) cryptography module which is woefully inadequate for today’s world will avoid many more headaches than getting rid of telnetlib et al (telnet!!!) will cause.
This is the most succinct explanation I’ve seen on how to start using smart pointers in C++, and when to use each.
Research Data Management and Analysis
Developing a modern data workflow for regularly updated data - Glenda M. Yenni et al, PLOS Biology
Updating Data Recipe - Ethan White, Albert Kim, and Glenda M. Yenni
This one’s a couple years old, and I’m surprised I hadn’t seen it before.
It’s getting easy to find good examples for scientists of getting started with GitHub, and then to CI/CD, for code. But for data it’s much harder. And there’s no reason why experimental data shouldn’t benefit from versioning, and analysis pipeline CI/CD that code does. As data gets cleaned up and the pipeline matures, and data products start being released, these tools are just as useful.
Here the authors publish a recipe, instructions, and template repos for Github Actions and Travis CI with data. The immediate audience is for ecology, but the process is pretty general. The instructions cover configuring the repo, connecting to Zenodo (for data artifacts) and the CI/CD tool. Then data checks can be added (with the pipeline failing if the data breaks a validity constraint). And data analysis can be done, with data products being published and versioned when a new release is created. It’s really cool!
By making the things we want to see (data checking, proper data releases) easier, as with automating them, we get more of what we want to see in the world. This is very useful work.
Jupyter Everywhere - Martin Renou, Jeremy Tuloup
We’ve talked about JupyterLite (#77, #112) before - a compiled-to-web-assembly JupterLite and Python engine that lets you run Jupyter entirely in the browser, with no aditional back end. Here Renou & Tuloup describes the latest version that has a REPL application that ships with default, so that the NumPy website lets you play with NumPy code interactively.
The authors walk you through several ways of “deploying” JupyterLite in static webpages, as a standalone static page in GitHub Pages or Vercel or Netlify, or including it in sphinx output for documentation, either as the repl or the entire notebook UI.
This is going to make teaching training sessions and giving demonstrations much easier, and make documentation much more interesting!
An Exploration of ‘Cloud-Native Vector’ - Chris Holmes
GeoParquet - Chris Holmes, Tom Augspurger, Joris Van den Bossche, Jésus Arroyo Torrens, Alberto Asuerro
I really like this and I don’t even do geospatial work.
In a lot of fields, file formats have grown in perceived importance well beyond any reasonable measure. (Hi, bioinformatics!) People spend a lot of time arguing how bytes should be arrayed on disk, rather than focussing what APIs the data libraries should support, pinning down the API, and then supporting one or more data layouts that support the APIs efficiently.
In geophysical data, NetCDF was a big help and nudged the community in productive directions. It laid out a pretty strict API for labelled gridded data, with a couple underlying file representations. Conventions (basically formats) developed around how to store different kinds of data in the NetCDF APIs, and that was that. File formats changed over time (including pretty drastically with NetCDF4), and it was all cool because there were tools to convert.
Parquet is becoming that lingua franca for columns of data, which is what you frequently want for reading data into memory or streaming through it to get relevant pieces. GeoParquet is a library (and proposed de facto standard) for storing GIS-type data in Parquet. One of the lovely things about Parquet is that it ties nicely into Arrow for computing on or interchanging the at a in memory.
In the article, Holmes lays out the advantages of Parquet format to some other attempts. He refers to Parquet (and some alternatives) as cloud-native, but efficient columnar data formats work just as well streaming through data on-prem, even on block storage. Parquet is built to support cloud-native things like streaming data, but that’s not necessary for this to be a useful format. The repository at the second link has v0.1 of the specification.
Research Computing Systems
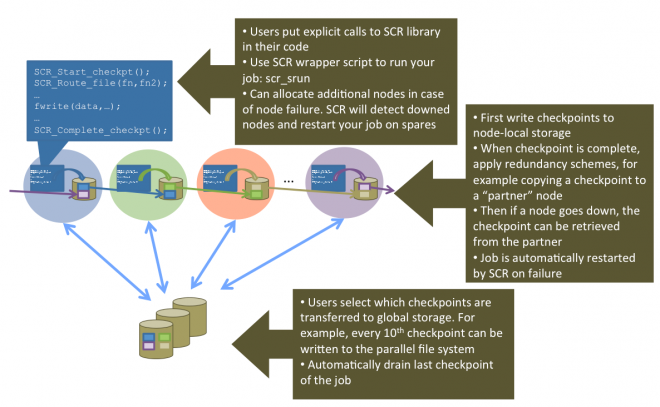
SCR: Scalable Checkpoint/Restart for MPI - LLNL
Coming back to HPC, I’ll probably be “discovering” things that have been going on for a while when I wasn’t paying attention - apologies in advance. This is the first I’ve heard of LLNL’s open source scalable checkpoint/restart library, which already had it’s v3.0 come out recently. It makes use of the increasingly hierarchical nature of storage, flushing checkpoints out to local storage, and then only periodically pushing one of the checkpoints out to global persistent storage.
cr8escape: New Vulnerability in CRI-O Container Engine Discovered by CrowdStrike (CVE-2022-0811) - John Walker & Manoj Ahuje, CloudStrike
Good overview by Walker & Ahuje of a vulnerability in the CRI-O container runtime, used by default in many new kubernetes installs, which lets users who can create pod definitions initiate pods which can be escaped from due to addition of support for (and insufficient validation of arguments to) sysctl.
Philosophically, I’m actually pleased to see that published exploits for escaping container runtimes are becoming increasingly either complicated or reliant on processes around operation; things are getting very mature. On a more practical nature, if you’re running CRI-O 1.19 and up, upgrade to patched versions, downgrade to 1.18, an/or add k8s policies to block pods that contain sysctl settings containing “+” or “=”.
Emerging Technologies and Practices
Automating OpenStack database backups with Kayobe and GitHub Actions - StackHPC
In most of research computing, gitops isn’t advanced much past software deveclopment CI/CD pipelines (which are already great!). But we can do a lot more tying version control changes to configuration files to deployment. Here the StackHPC team walks through using GitHub Actions (or GitLab CI/CD) to automating open stack database changes using their tool for Kayobe automation.
Growth in FLOPs used to train ML models - Derek Jones
Compute Trends Across Three Eras of Machine Learning - Jaime Sevilla, Lennart Heim, Anson Ho, Tamay Besiroglu, Marius Hobbhahn, Pablo Villalobos, arXiv:2202.05924
Jones has a nice summary of a paper by Sevilla et al., who went through the literature finding 493 ML models, and recording estimated flops for 129 of them, and very helpfully made their data available. Sevilla et al. looks at the data through the lens of pre-deep learning, the start of deep learning, and large-scale deep learning. Jones looks at it from the kinds of compute systems used: generic university clusters or other DIY systems, cloud computing, and more recently some special-purpose deep learning systems. It’s an interesting and fairly clear trifurcation:

Jones concludes with:
Supercomputer users have been facing the possibility of hitting the wall of maximum compute for over a decade. ML training is still a minnow in the supercomputer world, where calculations run for months, rather than a few days.
This overstates things in my opinion - recent peak usage of 1e24 FLOPs used to train a model is four months on a 100PF system assuming 100% efficiency - that’s a hero calculation by any measure. And usage is trending up very fast.
Random
Some war stories from that weird liminal time when the internet was starting to genuinely be a thing but dial-up modems were still key pieces of infrastructure. (SLIP! PPP! T1 Lines! Alternet!)
Ever want to design your own chip and have it fabbed in the dozens or few hundreds? That’s actually possible now.
Why typography is so hard on web pages. Bring back metal type!
A very deep dive, from one of the key authors, of the whats and whys of significant improvement of random number generation in Linux 5.17 and 5.18.
Another very deep dive, from people trying to reverse engineer it, into the Apple Silicon secure boot process. Through their efforts, you can now run Linux on your M1 Mac - Asahi Linux.
A little tool for building simple interacting systems (like predator-prey) and adding complexity, maybe useful for explaining simulations to school children? Loopy.
Writing scripts in golang.
Compiler tool to use machine learning to provide “profiling” information statically to link-time optimization steps.
Log and audit all SSH commands by using Cloudflare (or something else) as an SSH proxy.
Some problems in programming languages are very, very old. C++’s cstring.h strings vs std::string? Elixir’s binaries, strings, and charlists, or Erlang’s Strings/Binaries/IO Lists/IO Data? Rust’s String vs &str? New-fangled copycats. Meet COBOL’s two string types.
That’s it…
And that’s it for another week. Let me know what you thought, or if you have anything you’d like to share about the newsletter or management. Just email me or reply to this newsletter if you get it in your inbox.
Have a great weekend, and good luck in the coming week with your research computing team,
Jonathan
About This Newsletter
Research computing - the intertwined streams of software development, systems, data management and analysis - is much more than technology. It’s teams, it’s communities, it’s product management - it’s people. It’s also one of the most important ways we can be supporting science, scholarship, and R&D today.
So research computing teams are too important to research to be managed poorly. But no one teaches us how to be effective managers and leaders in academia. We have an advantage, though - working in research collaborations have taught us the advanced management skills, but not the basics.
This newsletter focusses on providing new and experienced research computing and data managers the tools they need to be good managers without the stress, and to help their teams achieve great results and grow their careers.
Jobs Leading Research Computing Teams
This week’s new-listing highlights are below; the full listing of 163 jobs is, as ever, available on the job board.
IT System Manager and Database Administrator - Oxford Universtiy, Nuffield Dept of Orthopaedics, Rheumatology, and Musculoskeletal Sciences, Oxford UK
We are seeking to appoint a highly qualified and dedicated IT System Manager and Database Administrator to join the research groups led by Professor Daniel Prieto-Alhambra at the Botnar Research Centre, Nuffield Department of Orthopaedics, Rheumatology and Musculoskeletal Sciences (NDORMS), Oxford. The Big Health Data Research group and the Pharmaco- and Device Epidemiology Research group are involved in a number of national and international studies. The former studies prevalent and rare conditions while the latter investigates the use and the risk-benefit of a number of licensed drugs, devices, and vaccines for the prevention and treatment of human disease in ‘real world’ conditions. As an IT System Manager and Database Administrator, reporting to an experienced computer scientist you will identify and lead highly technical IT projects from conception to completion defining the standards and making decisions that improve quality and efficiency of data harmonisation, curation, and processing within the Department.
Senior Front End Software Developer - Health Data Research UK, remote UK
HDR UK requires the services of an experienced senior developer to work on the next stage of its flagship Innovation Gateway and associated software services. The Innovation Gateway was first released in May 2020 and provides a key resource to discover and access health data resources for researchers and data custodians. Its primary function is providing visibility to and supporting access requests for data sets by researchers in the UK and around the globe to health data that can support them in their quest to deliver scientific insights, that improve health and healthcare of the UK and worldwide populations. The successful candidate will be expected to work within the Gateway agile software development team, providing development leadership and hands on implementation of key functionality and new concepts, primarily focused on the front-end aspects of the product.
Program Manager 2, HPC Workflows - Microsoft Azure, Redmond WA USA
As Azure’s portfolio of solutions for scalable HPC and AI workloads expands and evolves, we are looking for a program manager to help lead our current and next generation of HPC cluster and supercomputer VM products for HPC workloads (e.g. CFD, FEA, molecular dynamics, cryptanalytics, genomics, proteomics, risk analysis, rendering, high energy physics, energy research and exploration, weather and climate modeling, chemistry, quantum chromodynamics, high performance data analytics/machine learning, etc.) You will be responsible for defining product requirements based on input from HPC customers and practitioners, Azure engineering teams, and your own views, and distilling these perspectives into a coherent set of targets for Azure to address in its roadmaps and execution.
Senior Program Manager, HPC & AI, FPGA Instances - Microsoft Azure, Redmond WA USA
As Azure’s portfolio of solutions for scalable HPC and AI workloads expands and evolves, we are looking for a program manager to help lead our current and next generation of FPGA accelerated VM products for programmable hardware solutions. You will be responsible for defining product requirements based on input from FPGA compute customers and practitioners, Azure engineering teams, and your own views, and distilling these perspectives into a coherent set of targets for Azure to address in its roadmaps and execution.
You will also help define Azure’s broader strategy for FPGA compute products.
Computational Science Lead - Sanofi, Toronto ON CA or Boston MA USA or Barcelona ES
You are a dynamic data scientist interested in leading a group of ML and software engineers to develop and deploy advanced ML solutions that will change the way biomedical research is performed. We are looking for individuals ready to challenge the status quo to ensure development and impact of Sanofi’s AI solutions for developing the drugs and improving the clinical trials that will benefit the patients of tomorrow. You have a keen eye for improvement opportunities and a demonstrated ability to deliver AI/ML solutions while working across different technologies and in a cross-functional environment.
Director, Organizational Research IT - National Research Council, various CA
The Director, Organizational Research IT is accountable for ensuring that NRC research is effectively enabled by information technology (IT) infrastructure, Information Management (IM) Data, tools, and services. This includes managing all IT/IM and Data elements of the KITS digital pillar targeting Excellence in Research and Innovation, including providing secretariat support for digital research governance, translating governance decisions into action, maintaining an effective self-serve cloud capability in support of NRC research, coordinating research IT project intake, providing leadership for optimizing use of research software across the NRC, collaborating closely with external and internal partners to meet research clients’ IM/IT and data requirements, and providing leadership and vision for planning the continuous improvement and evolution of the research IT service suite of tools and services.
Data Science Senior Manager - Diageo, Edinburgh or Glasgow or London UK or Budapest HU
Build Data science solutions for business functions to improve business growth and efficiency. Manage the activity of data science teams collaborating with A&I leaders and business customers. Execute data science projects using a range of datasets and techniques. Work with senior stakeholders and market leads to develop the Proof Of Concept (POC) and then scale it for other regions. Ensure that the solution produces insights that the business can interpret and execute on
Scientific Data Management Group Leader - Evotec, Abingdon UK
As part of the Global Research Informatics, Scientific Data Management (SDM) department, the SDM Group Leader will lead the team in implementing Drug Discovery and Development applications, coordinate local and global Informatics projects, and manage data for customers’ scientific projects. Reporting to our VP Head of Scientific Data Management, you will be an experienced line manager of a group of talented information scientists, and will develop new skills and competencies to drive the informatics transformation at Evotec.
Lead HPC (High Performance Computing) and Research Computing Engineer - University of Nevada, Reno, Reno NV USA
The Office of Information Technology at the University of Nevada, Reno is seeking a Lead HPC (High Performance Computing) and Research Computing Engineer to advance, architect, and manage the HPC and related research technology ecosystem at our Carnegie R1 University. Reporting to the Director of Cyberinfrastructure, this position collaborates with personnel across central and distributed IT to manage and deliver timely and appropriate research computing and data (RCD) platforms and services to faculty and students. The Lead HPC and Research Computing Engineer is responsible for leading design, implementation, and support of the University’s central research computing infrastructure. This includes HPC clusters, interactive computing hosts, large-scale storage systems, advanced networks, and associated management and monitoring infrastructure. The ideal candidate’s primary motivation should be to align systems design, workflows, and team effort to maximize research outcomes and user experience across a wide range of science and engineering domains. The measures of success for the campus Cyberinfrastructure team are if time-to-science is reduced and if a wide range of researchers/students have positive experiences and are successful in using our technologies.
Research Computing Supervisor - Sunnybrook Health Sciences Centre Research Institute, Toronto ON CA
The Research Computing Supervisor is responsible for managing SRI computing deskside operations and supporting corporate medium to large enterprise wide projects as assigned by the corporate IT group (CS/TS/Infosecurity).
Senior Data Science Delivery Manager - UK Office for National Statistics, Darlington or Newport or Fareham UK
The Data Science Campus, part of the Office for National Statistics, is a purpose-built centre of excellence for data science. We are the hub of the UK’s public sector data science landscape and play a leading role in developing the UK’s capability and international reputation in data science. A Senior delivery manager is accountable for the end-to-end delivery of innovative research, methods development, products, and services.
Scientific Manager - UK Met Office, Exeter UK
The observations R&D team develops improvements to our observations from inception to delivery into operations across many systems including radar, remote sensing and surface-based observations. The team is multi-disciplinary consisting of engineers, software developers and scientists - all working towards a common goal. It is focused on delivering improved observations across a wide range of environmental measurement systems including land and marine automatic weather stations and optically based measurement systems
Manager Research Project B (Clinical Research Computing Unit) - University of Pennsylvania, Philadelphia PA USA
The Clinical Research Computing Unit (CRCU) is an Academic Clinical Research Organization within the Center for Clinical Epidemiology and Biostatistics (CCEB) in the Perelman School of Medicine at the University of Pennsylvania. Since its inception in 1997, the CRCU has been expertly providing the full range of services essential for the conduct of clinical research projects, including Phase I-IV, multi-center, randomized, clinical trials, registry, and cohort studies utilizing state-of-the-art technology and tools to ensure superior data quality. The CRCU provides expertise in project management, data coordination and research computing tailored to meet your project requirements. The CRCU project teams’ partner with the Biostatistics Analysis Center (BAC) in the project design phase to plan accurate and precise data collection modules and to structure project reports for steady oversight. We specialize in study design and development, site management and training, data collection, processing, quality control, regulatory requirements and reporting, database development, administration, security, data storage and proposal development.
Project Manager for Research Computing - Harvard Medical School, Boston MA USA
Specializing in running projects for IT in research lab environments, the Project Manager will guide and plan for project success from project inception through project completion. The project manager will lead the stakeholder communications program; coordinating directly with functional process and data owners to understand and incorporate multiple functions and user perspectives into the project and plan for successful user acceptance testing and rollout to meet end user needs and school objectives. The project manager will seek out and coordinate with other IT leaders and project managers, technologists, researchers, and support staff to ensure projects are planned for successful transition to production support, and to improve the maturity of the IT Project Management processes and toolsets for enhanced collaboration across IT and HMS. This position will have a broad scope and impact across Research Computing and the Information Technology department; the incumbent will focus on developing strong business partnerships and cultivating trust.
Head of Clinical and Research Data - University College London Hospitals, London UK
We are creating a new team with the mission to put high quality datasets into the hands of clinical and research teams in a scalable, efficient manner, helping UCLH advance research and deliver top-quality patient care The successful applicant will lead this team and will coordinate delivery of multiple. ongoing projects through the development of high-quality data models and data pipelines. We are looking for an ambitious, motivated, and dynamic candidate with experience managing complex data / software projects including their requirements, agile delivery, communication, and staffing. They will work closely with the architects in the organization to ensure that the implementation is accurate, secure, scalable, and maintainable. The resulting system will provide a highly usable, consolidated view of our datasets and business rules and enable self-service business intelligence and analytics.
Data Science Leader - Anaconda, remote US
Anaconda is seeking people who want to play a role in shaping the future of enterprise machine learning, and data science. Candidates should be knowledgeable and capable, but always eager to learn more and to teach others. Overall, we strive to create a culture of ability and humility and an environment that is both relaxed and focused. We stress empathy and collaboration with our customers, open-source users, and each other. Anaconda is seeking a talented Data Science Leader to join our rapidly-growing company. This is an excellent opportunity for you to leverage your experience and skills and apply it to the world of data science and machine learning.
Scientific Computing Senior Manager - Fred Hutchinson Cancer Center, Seattle WA USA
The Scientific Computing Senior Manager will direct and oversee all facets of scientific computing including managing a highly capable engineer team, engineering functions such as design, development, installation, and maintenance of hardware and software, and customer service and support, for the organization. The senior manager will create and oversee partnerships with faculty and stakeholders in the scientific community to test and integrate new analysis pipelines, determine innovative technologies and software to support their research, evangelize new and current solutions, and foster technology and data storage best practices. The senior manager will participate in strategic planning and align projects and resources for implementation in partnership with the Project Management Office and Business Operations, and other departments.